removing noise using connected components
Hi all,
I am running this in OpenCV 3.x and Python
I have an image like this: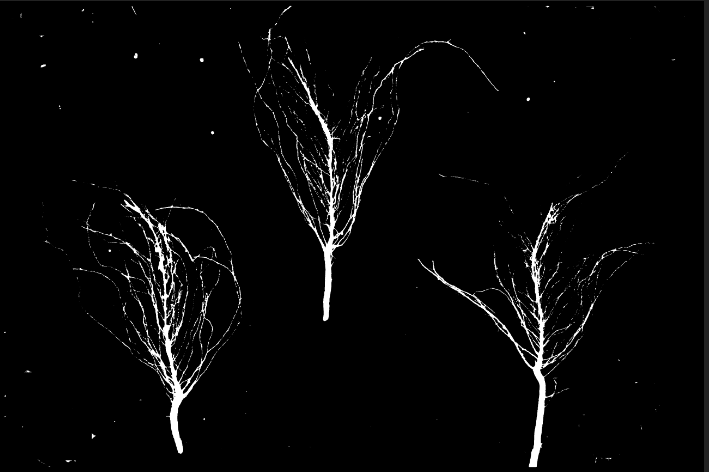 Its an
HSV thresholded output of a BGR image. I am trying to crop the roots alone. I have been able to successfully do dilation and erosion, then contour detection on certain images. But on others, while dilating the image, I have a lot of unwanted small white blobs which get dilated along with the other pixels and cause trouble when identifying the contours.
Its an
HSV thresholded output of a BGR image. I am trying to crop the roots alone. I have been able to successfully do dilation and erosion, then contour detection on certain images. But on others, while dilating the image, I have a lot of unwanted small white blobs which get dilated along with the other pixels and cause trouble when identifying the contours.
Normal morphological methods like blurring, eroding also destroy my root pixels which render the image useless.
So basically I want to selectively target the other specks and fill them black so I can dilate better.
I have also tried connected components on opencv, converted the labels in terms of hue and got this:
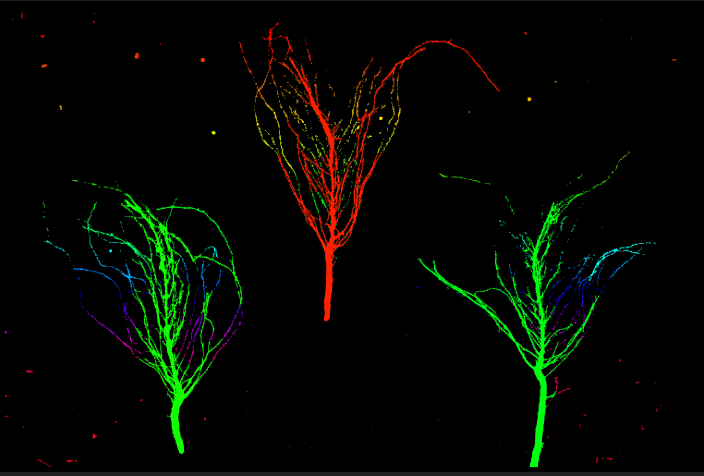
Here is my code:code:
img = cv2.imread('D:/plants/kevin/hsv.jpg',0)
nlabel,labels,stats,centroids = cv2.connectedComponentsWithStats(img,connectivity=8)
img = cv2.imread('D:/plants/kevin/hsv.jpg',0)
nlabel,labels,stats,centroids = cv2.connectedComponentsWithStats(img,connectivity=8)
for l in range(1,nlabel):
if stats[l,cv2.CC_STAT_AREA]<=1000:
labels_small.append(l)
areas_small.append(stats[l,cv2.CC_STAT_AREA])
mask = np.ones_like(labels,dtype=np.uint8)
tstart = time.time()
for l in labels_small:
mask[labels == l] = 0
#img[labels == l] = 0
tend = time.time()
print 'time taken:' + str(tend-tstart) + 'secs'
im = Image.fromarray(mask)
im.convert('RGB').save("D:/plantcv2/kevin/cc_mask.jpg")
newHsv = copy.deepcopy(img)
cv2.bitwise_and(newHsv,newHsv,mask=mask)
cv2.imwrite('D:/plants/kevin/hsv_new.jpg',newHsv)
Is it possible to identify the connected components which have the least area and not part of the root and flood fill them black? Let me know what approaches would work. Thanks ! Thanks! :)
 Its an
HSV thresholded output of a BGR image. I am trying to crop the roots alone. I have been able to successfully do dilation and erosion, then contour detection on certain images. But on others, while dilating the image, I have a lot of unwanted small white blobs which get dilated along with the other pixels and cause trouble when identifying the contours.
Its an
HSV thresholded output of a BGR image. I am trying to crop the roots alone. I have been able to successfully do dilation and erosion, then contour detection on certain images. But on others, while dilating the image, I have a lot of unwanted small white blobs which get dilated along with the other pixels and cause trouble when identifying the contours.

