MSER and SURF provide false matches
After using MSER as feature detection algorithm and SURF as descriptor extractor algorithm, i got the below image and i think it represents false matchings. please have alook at the picture and notice the green lines and the pink one. these lines are linked to different features, because i assume that these lines produced from the matching process should be linked to the same features.
please let me know why i am getting false matches and how to solve this issue
Note:
I used also ORB algorithm as for feature detection and descriptor extractor and i received the first image posted below without aplying any geometric transformation/correction using RANSAC.
does it means that ORB algorithm does not require applying RANSAC?
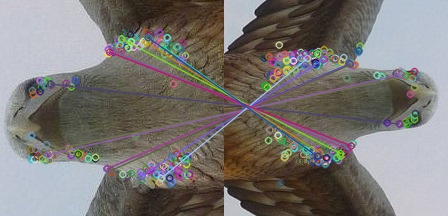
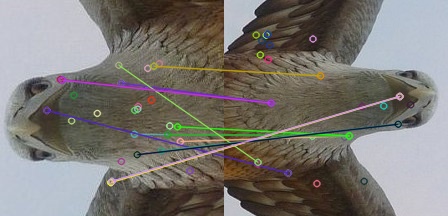

You will need to apply some sort of outlier detection like RANSAC, which will simply discard those matches.
@StevenPuttemans please see the note i posted above.
can you please provide an example of how to do the RANSAC? and should i do the RANSAC on the image i got from the descriptor matcher? also is there any recommednded algorithms for features detection, descriptor extactor and descriptor matcher?
The documentation has a very nice example of findHomography using RANSAC
@dirtbag thanks fo rthe reference. i just have a question please, after the matching process, why do i need to make a threshold while i can just sort the matches yielded from the .match(.....) method ascendinly, and then pich the top 20 or 10 ascendingly sorted distances?
kind regards,
It's mostly to save computation time during RANSAC. If you throw out a bunch of bad matches using a simple distance threshold (3*min_dist), then findHomography has a better chance of finding the right homography quickly. That said, you're idea of only keeping the best K matches would accomplish the same goal of filtering/shortening the list of matches.
Keep in mind that there are images where several keypoints will match perfectly with many, many other keypoints. In this case, you'll need a larger list to be sure that you'll find a good solution. 3*min_dist would then give you the larger list you need while filtering out at least some of the garbage.