Detecting overlapping circles
Hi all,
This is my first post in this forum, and it could be that i'm totally in the wrong place. But i give it a shot..
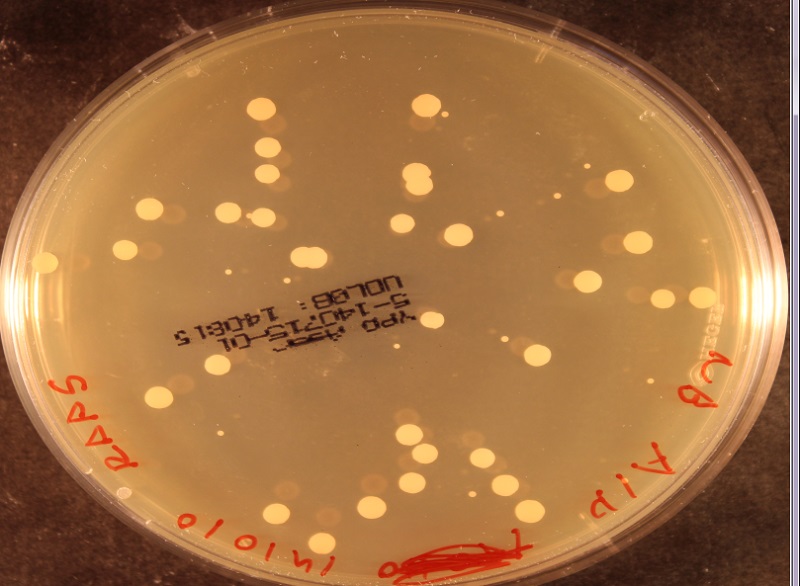
I'm a biologist tasked with detecting yeast colonies on a nutrient plate. The colonies are circular and about 3-5 mm in diameter. My problem comes from that several colonies might overlap so that you have shapes consisting of 2-5 partially overlapping circles. I need to detect the position and size of all such circles.
I know nothing about computer vision/pattern recognition, but this is my naive approach so far:
1) Find the edges of the shapes. I do this by calculating the color variance for each pixel and its surrounding neighbours. This gives a map where edges have high values and the background has zero (after some filtering)
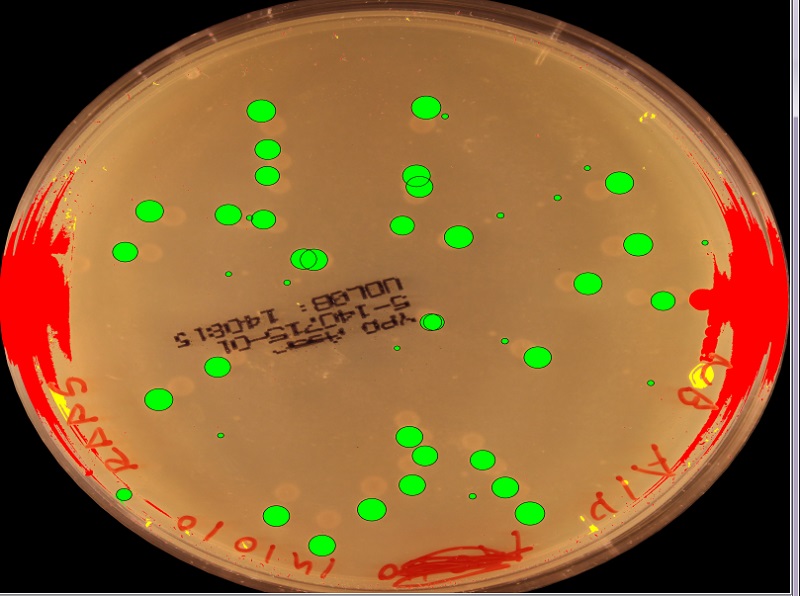
2) Find all the individual shapes (connected non-zero regions in the map above)
3) a) For each shape select a random connected subset of the shape (corresponding to a partial circle segment). Fit a circle to these points by least squares. b) Repeat a) several times to get coverage for the whole edge of the shape. c) Cluster the calculated circle geometries from above. The number of clusters should then give the number of overlapping circles and the mean och each cluster should give the geometry.
4) (Optional. Use the input from 3) to perform a constrained heuristic optimization to fine tune the fittings.)
This whole thing works reasonably well if the circles are few and only slightly overlapping, but not well for complex shapes with several circles. It's also slow because of the fitting of random arcs. It also feels like too much of a home made brute force solution and i'm sure there are many more clever ways to do it. For example, could you count the number of times some fitted shape (spline or something) goes from concave to convex as you go along the edge of the shape? That should give the number of circles and their approximate intersection points.
All of this is done in Matlab, but anything else would work as well. Here is an image which shows how the colonies might look: http://medicine.emory.edu/id/labs/lyon/yeast.bmp
Any help is very welcome! Feeling rather lost in this field :)

Have you tried the hough_circles? Or a kind of binarization and then fitellipse; if it is too elliptic/long, then there are more circles (approximatively hi_ray/lo_ray)?
Thanks for your suggestions! It was not stated in the original question, but I actually use an elliptical fit just as you suggested in order to screen out the shapes which are simple circles. The above workflow is only applied to complex shapes.
I looked up Hough transform at Wikipedia, and it sounds rather similar to what I do. I take a random segment, fit a circle to it, clusters in parameter space and use the median in each cluster as representative. Hough transform iterates over each pixel, fits a circle to it and its surrounding and counts how ofter the resulting parameter combination occurs. The main difference seems to be that Hough does this for each pixel while I do it for some random number of segments. The counting also indicates that Hough requires some bounds on the...
...parameter space. If so then my problem can maybe be reduced to how to fit circles to a small number of points. The reason I use larg(er) random segments is that the quadratic fitting tends to favor very small circles when only a small number of pixels are used. Is there maybe a clever way of doing this? I see unpleasent words such as eigenvectors coming up.. :)
Sorry, small number of closely aligned points I mean. So that only some small fraction of the circle is actually covered.
Have a look on Morphology Transformations, you need to proceed like, threshold->morphology->contour analysis etc...