Negative values in filter2D convolution
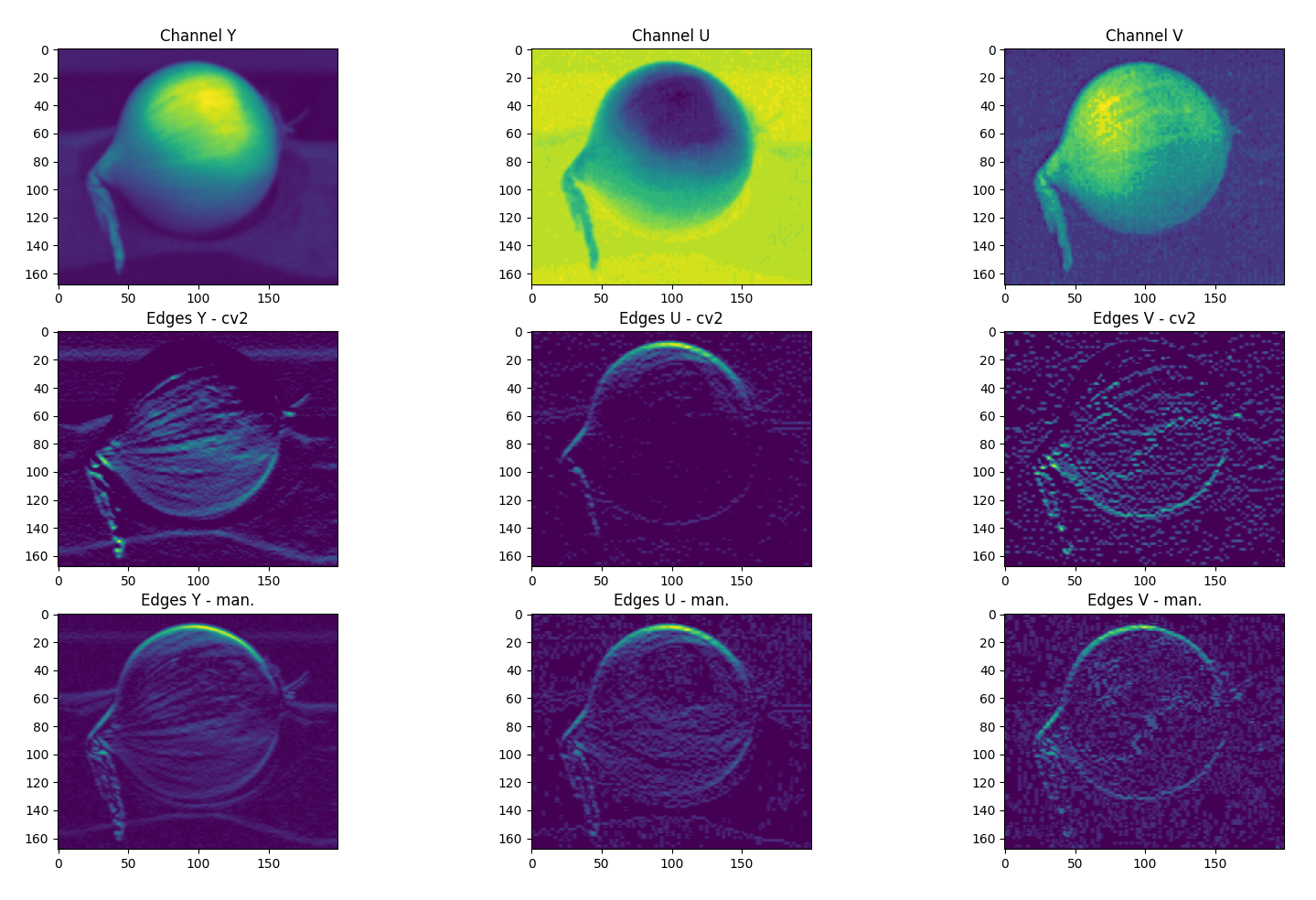
I am playing around with an edge detection algorithm on a .JPG image using OpenCV in Python. For comparison I also created a simple convolution function that slides a kernel over every pixel in the image. However, the results between this 'manual' convolution and cv2::filter2D are quite different as can be seen in the attached picture. The filter2D function (middle row) is very fast compared to my implementation (bottom row) but the results miss some edges. In the attached example, a simple 3x3 horizontal ([121][000][-1-2-1]) kernel is used and we can already see notable differences.
This seems to be caused by the filter2D function surpressing negative values. When I flip the kernel, the missing edges are displayed, so the image is equal to the difference between the images in row 2 (cv2) and row 3 (my implementation). Is there an option that allows filter2D to also record negative (or absolute magnitude) values? A simple workaround would be to do convolutions for the kernel and the 180 degrees flipped version and sum the two results, but this will unnecessarily complicate the code.
Since I have little ambition to rewrite the filter2D function to suit my needs (since my simple implementation is too slow), I wondered whether there is an official workaround? Regardless of a solution, I think it would be good to update the documentation.
A minimal example using a .JPG image img with kernel as described above would be:
imgYUV = cv2.cvtColor(img, cv2.COLOR_BGR2YUV)
clr1, clr2, clr3 = cv2.split(imgYUV)
pad=1
paddedImg = cv2.copyMakeBorder(clr1, pad,pad,pad,pad,cv2.BORDER_REPLICATE)
(iH, iW) = paddedImg.shape[:2]
resultsMyImplementation = np.zeros((iH,iW,1), dtype='int32')
for y in np.arange(pad, iH+pad):
for x in np.arange(pad, iW+pad):
roi = paddedImg[y-pad,y+pad+1, x-pad:x+pad+1]
k = (roi*kernel).sum()
resultsMyImplementation[y-pad,x-pad] = k
resultsCV2 = cv2.filter2D(clr1, -1, kernel)

show code, please.
Added code for clarity.
have a look at the docs again, your implementation differs
what is the type of
img?The image is a JPG, updated code to reflect conversion to np.ndarray() containing only 1 color channel. Results for all 3 channels can be seen in the image.