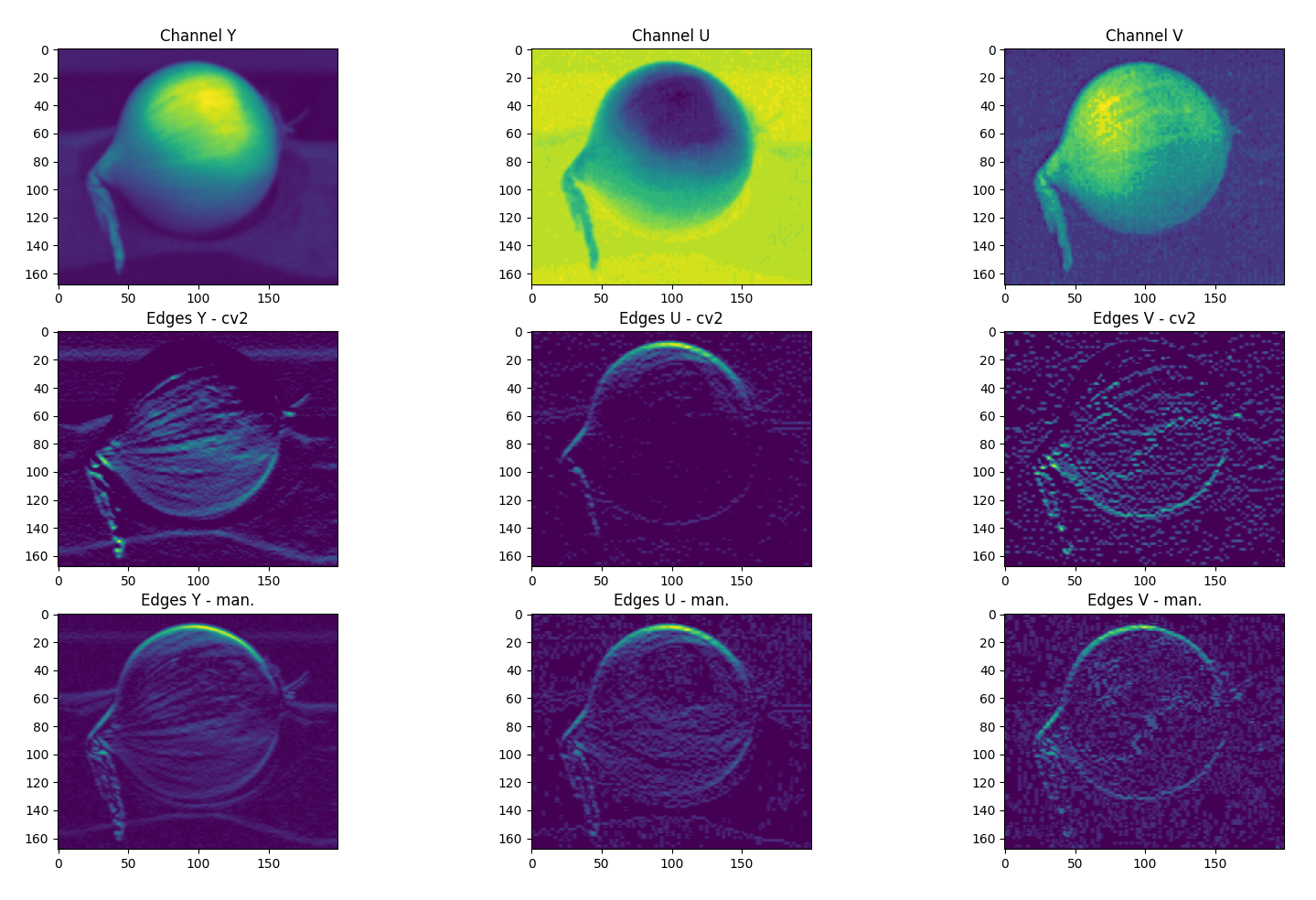
I am playing around with an edge detection algorithm using OpenCV in Python. For comparison I also created a simple convolution function that slides a kernel over every pixel in the image. However, the results between this 'manual' convolution and cv2::filter2D are quite different as can be seen in the attached picture. The filter2D function (middle row) is very fast compared to my implementation (bottom row) but the results miss edges. In the attached example, a simple 3x3 horizontal ([121][000][-1-2-1]) kernel is used and we can already see notable differences. This might be a possible result of filter2D flipping the kernel. However, I save the input as abs(cv2.filter2D(img, -1, kernel)) and abs((roi*kernel).sum()) to make sure no negative values are present.
Because of the results, I suspect that filter2D does some filtering or other image processing, either before or after applying the convolution. It's also possible that negative values are set to zero. I have little ambition to rewrite the whole filter2D function (since my simple implementation is too slow) and wondered whether it is possible to turn this off and if so, can the documentation be updated?