This forum is disabled, please visit https://forum.opencv.org
 | 1 | initial version |
I tried your program. I think there is a problem when you initialize the size of cm1 and cm2 (it should be [3][3]):
double cm1[3][1] = {{8.941981e+02, 0.000000e+00, 6.601406e+02}, {0.000000e+00, 8.927151e+02, 2.611004e+02}, {0.000000e+00, 0.000000e+00, 1.000000e+00}}; double cm2[3][2] = {{8.800704e+02, 0.000000e+00, 6.635881e+02 }, {0.000000e+00, 8.798504e+02, 2.690108e+02}, {0.000000e+00, 0.000000e+00, 1.000000e+00}};
It tried to visualize your result with Meshlab (rename the file to .xyz and remove the header):

For
reprojectImageTo3D
I think that it is better to use the raw disparity instead of the normalize one. The result:

I don't think there is a problem when you reconstruct the 3D the point cloud.
 | 2 | No.2 Revision |
I tried your program. I think there is a problem when you initialize the size of cm1 and cm2 (it should be [3][3]):
double cm1[3][1] = {{8.941981e+02, 0.000000e+00, 6.601406e+02}, {0.000000e+00, 8.927151e+02, 2.611004e+02}, {0.000000e+00, 0.000000e+00, 1.000000e+00}}; double cm2[3][2] = {{8.800704e+02, 0.000000e+00, 6.635881e+02 }, {0.000000e+00, 8.798504e+02, 2.690108e+02}, {0.000000e+00, 0.000000e+00, 1.000000e+00}};
It tried to visualize your result with Meshlab (rename the file to .xyz and remove the header):

For
reprojectImageTo3D
I think that it is better to use the raw disparity instead of the normalize one. The result:

I don't think there is a problem when you reconstruct the 3D the point cloud.
Edit:
When I load the point cloud (your pcd file renamed in xyz and without the header part) into Meshlab, I get this:
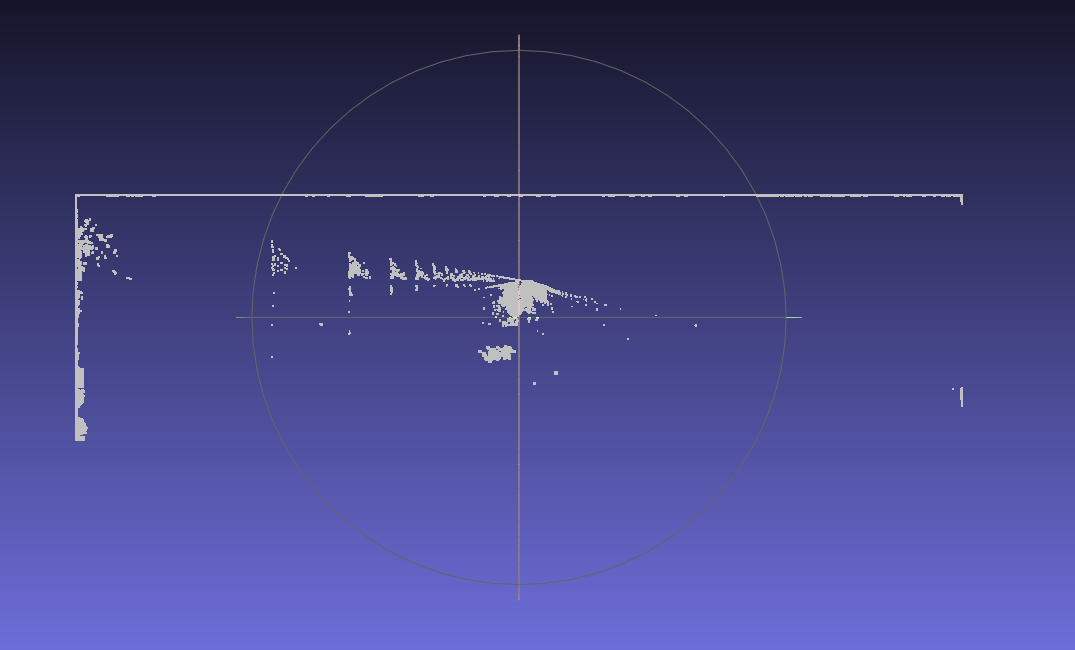
and I have to move around to get the view of the first image (rotation on two axis + zoom).
Also, to obtain the point cloud in the second image, I had to manually threshold the value:
if(points.at<Vec3f>(i,j)[2] < 10) {
//Write
}
I use this tutorial to get better visualization.
The code I used:
#include <iostream>
#include <fstream>
#include <opencv2/core/core.hpp>
#include <opencv2/calib3d/calib3d.hpp>
#include <opencv2/highgui/highgui.hpp>
#include <opencv2/imgproc/imgproc.hpp>
using namespace cv;
using namespace std;
int main()
{
Mat img1, img2;
img1 = imread("left.png", CV_LOAD_IMAGE_GRAYSCALE);
img2 = imread("right.png", CV_LOAD_IMAGE_GRAYSCALE);
double cm1[3][3] = {{8.941981e+02, 0.000000e+00, 6.601406e+02}, {0.000000e+00, 8.927151e+02, 2.611004e+02}, {0.000000e+00, 0.000000e+00, 1.000000e+00}};
double cm2[3][3] = {{8.800704e+02, 0.000000e+00, 6.635881e+02 }, {0.000000e+00, 8.798504e+02, 2.690108e+02}, {0.000000e+00, 0.000000e+00, 1.000000e+00}};
double d1[1][5] = {{ -3.695739e-01, 1.726456e-01, -1.281525e-03, 1.188796e-03, -4.284730e-02}};
double d2[1][5] = {{-3.753454e-01, 1.843265e-01, -1.307069e-03, 2.190397e-03, -4.989103e-02}};
Mat CM1 (3,3, CV_64FC1, cm1);
Mat CM2 (3,3, CV_64FC1, cm2);
Mat D1(1,5, CV_64FC1, d1);
Mat D2(1,5, CV_64FC1, d2);
cout << "Calibration matrix left:\n" << CM1 << endl;
cout << "Distorstion matrix left:\n" << D1 << endl;
cout << "Calibration matrix right:\n" << CM2 << endl;
cout << "Distorstion matrix right:\n" << D2 << endl;
double r[3][3] = {{9.998381e-01, 1.610234e-02, 8.033237e-03},{-1.588968e-02, 9.995390e-01, -2.586908e-02 },{-8.446087e-03, 2.573724e-02, 9.996331e-01}};
double t[3][4] = {{ -5.706425e-01}, {8.447320e-03}, {1.235975e-02}};
Mat R (3,3, CV_64FC1, r);
Mat T (3,1, CV_64FC1, t);
//Mat R, T;
Mat R1, R2, T1, T2, Q, P1, P2;
stereoRectify(CM1, D1, CM2, D2, img1.size(), R, T, R1, R2, P1, P2, Q);
Mat map11, map12, map21, map22;
Size img_size = img1.size();
initUndistortRectifyMap(CM1, D1, R1, P1, img_size, CV_16SC2, map11, map12);
initUndistortRectifyMap(CM2, D2, R2, P2, img_size, CV_16SC2, map21, map22);
Mat img1r, img2r;
remap(img1, img1r, map11, map12, INTER_LINEAR);
remap(img2, img2r, map21, map22, INTER_LINEAR);
//img1 = img1r;
//img2 = img2r;
int sadSize = 3;
StereoSGBM sbm;
sbm.SADWindowSize = sadSize;
sbm.numberOfDisparities = 144;//144; 128
sbm.preFilterCap = 10; //63
sbm.minDisparity = 0; //-39; 0
sbm.uniquenessRatio = 10;
sbm.speckleWindowSize = 100;
sbm.speckleRange = 32;
sbm.disp12MaxDiff = 1;
sbm.fullDP = true;
sbm.P1 = sadSize*sadSize*4;
sbm.P2 = sadSize*sadSize*32;
Mat disp, disp8;
sbm(img1, img2, disp);
normalize(disp, disp8, 0, 255, CV_MINMAX, CV_8U);
Mat points, points1;
reprojectImageTo3D(disp, points, Q, true);
cvtColor(points, points1, CV_BGR2GRAY);
ofstream point_cloud_file;
point_cloud_file.open ("point_cloud.xyz");
for(int i = 0; i < points.rows; i++) {
for(int j = 0; j < points.cols; j++) {
if(points.at<Vec3f>(i,j)[2] < 10) {
point_cloud_file << points.at<Vec3f>(i,j)[0] << " " << points.at<Vec3f>(i,j)[1] << " " << points.at<Vec3f>(i,j)[2]
<< " " << static_cast<unsigned>(img1.at<uchar>(i,j)) << " " << static_cast<unsigned>(img1.at<uchar>(i,j)) << " " << static_cast<unsigned>(img1.at<uchar>(i,j)) << endl;
}
}
}
point_cloud_file.close();
imshow("Img1", img1);
imshow("Img2", img2);
imshow("points", points);
imshow("points1", points1);
//imwrite("disparity.jpg", disp8);
//imwrite("points1.jpg", points1);
//imwrite("points.jpg", points);
//for(int i=0; i<points.rows; ++i)
//{
// Point3f* point = points.ptr<Point3f>(i) ;
// for(int j=0; j<points.cols; ++j)
// {
// out<<i<<" "<<j<<" x: "<<(*point).x<<" y: "<<(*point).y<<" z: "<<(*point).z<<endl;
// ++point;
// }
//}
waitKey(0);
return 0;
}
 | 3 | No.3 Revision |
I tried your program. I think there is a problem when you initialize the size of cm1 and cm2 (it should be [3][3]):
double cm1[3][1] = {{8.941981e+02, 0.000000e+00, 6.601406e+02}, {0.000000e+00, 8.927151e+02, 2.611004e+02}, {0.000000e+00, 0.000000e+00, 1.000000e+00}}; double cm2[3][2] = {{8.800704e+02, 0.000000e+00, 6.635881e+02 }, {0.000000e+00, 8.798504e+02, 2.690108e+02}, {0.000000e+00, 0.000000e+00, 1.000000e+00}};
It tried to visualize your result with Meshlab (rename the file to .xyz and remove the header):header) as I don't have for the moment PCL:

For
reprojectImageTo3D
I think that it is better to use the raw disparity instead of the normalize one. The result:

I don't think there is a problem when you reconstruct the 3D the point cloud.
Edit:
When I load the point cloud (your pcd file renamed in xyz and without the header part) into Meshlab, I get this:

and I have to move around to get the view of the first image (rotation on two axis + zoom).
Also, to obtain the point cloud in the second image, I had to manually threshold the value:
if(points.at<Vec3f>(i,j)[2] < 10) {
//Write
}
I use this tutorial to get better visualization.
The code I used:
#include <iostream>
#include <fstream>
#include <opencv2/core/core.hpp>
#include <opencv2/calib3d/calib3d.hpp>
#include <opencv2/highgui/highgui.hpp>
#include <opencv2/imgproc/imgproc.hpp>
using namespace cv;
using namespace std;
int main()
{
Mat img1, img2;
img1 = imread("left.png", CV_LOAD_IMAGE_GRAYSCALE);
img2 = imread("right.png", CV_LOAD_IMAGE_GRAYSCALE);
double cm1[3][3] = {{8.941981e+02, 0.000000e+00, 6.601406e+02}, {0.000000e+00, 8.927151e+02, 2.611004e+02}, {0.000000e+00, 0.000000e+00, 1.000000e+00}};
double cm2[3][3] = {{8.800704e+02, 0.000000e+00, 6.635881e+02 }, {0.000000e+00, 8.798504e+02, 2.690108e+02}, {0.000000e+00, 0.000000e+00, 1.000000e+00}};
double d1[1][5] = {{ -3.695739e-01, 1.726456e-01, -1.281525e-03, 1.188796e-03, -4.284730e-02}};
double d2[1][5] = {{-3.753454e-01, 1.843265e-01, -1.307069e-03, 2.190397e-03, -4.989103e-02}};
Mat CM1 (3,3, CV_64FC1, cm1);
Mat CM2 (3,3, CV_64FC1, cm2);
Mat D1(1,5, CV_64FC1, d1);
Mat D2(1,5, CV_64FC1, d2);
cout << "Calibration matrix left:\n" << CM1 << endl;
cout << "Distorstion matrix left:\n" << D1 << endl;
cout << "Calibration matrix right:\n" << CM2 << endl;
cout << "Distorstion matrix right:\n" << D2 << endl;
double r[3][3] = {{9.998381e-01, 1.610234e-02, 8.033237e-03},{-1.588968e-02, 9.995390e-01, -2.586908e-02 },{-8.446087e-03, 2.573724e-02, 9.996331e-01}};
double t[3][4] = {{ -5.706425e-01}, {8.447320e-03}, {1.235975e-02}};
Mat R (3,3, CV_64FC1, r);
Mat T (3,1, CV_64FC1, t);
//Mat R, T;
Mat R1, R2, T1, T2, Q, P1, P2;
stereoRectify(CM1, D1, CM2, D2, img1.size(), R, T, R1, R2, P1, P2, Q);
Mat map11, map12, map21, map22;
Size img_size = img1.size();
initUndistortRectifyMap(CM1, D1, R1, P1, img_size, CV_16SC2, map11, map12);
initUndistortRectifyMap(CM2, D2, R2, P2, img_size, CV_16SC2, map21, map22);
Mat img1r, img2r;
remap(img1, img1r, map11, map12, INTER_LINEAR);
remap(img2, img2r, map21, map22, INTER_LINEAR);
//img1 = img1r;
//img2 = img2r;
int sadSize = 3;
StereoSGBM sbm;
sbm.SADWindowSize = sadSize;
sbm.numberOfDisparities = 144;//144; 128
sbm.preFilterCap = 10; //63
sbm.minDisparity = 0; //-39; 0
sbm.uniquenessRatio = 10;
sbm.speckleWindowSize = 100;
sbm.speckleRange = 32;
sbm.disp12MaxDiff = 1;
sbm.fullDP = true;
sbm.P1 = sadSize*sadSize*4;
sbm.P2 = sadSize*sadSize*32;
Mat disp, disp8;
sbm(img1, img2, disp);
normalize(disp, disp8, 0, 255, CV_MINMAX, CV_8U);
Mat points, points1;
reprojectImageTo3D(disp, points, Q, true);
cvtColor(points, points1, CV_BGR2GRAY);
ofstream point_cloud_file;
point_cloud_file.open ("point_cloud.xyz");
for(int i = 0; i < points.rows; i++) {
for(int j = 0; j < points.cols; j++) {
if(points.at<Vec3f>(i,j)[2] < 10) {
point_cloud_file << points.at<Vec3f>(i,j)[0] << " " << points.at<Vec3f>(i,j)[1] << " " << points.at<Vec3f>(i,j)[2]
<< " " << static_cast<unsigned>(img1.at<uchar>(i,j)) << " " << static_cast<unsigned>(img1.at<uchar>(i,j)) << " " << static_cast<unsigned>(img1.at<uchar>(i,j)) << endl;
}
}
}
point_cloud_file.close();
imshow("Img1", img1);
imshow("Img2", img2);
imshow("points", points);
imshow("points1", points1);
//imwrite("disparity.jpg", disp8);
//imwrite("points1.jpg", points1);
//imwrite("points.jpg", points);
//for(int i=0; i<points.rows; ++i)
//{
// Point3f* point = points.ptr<Point3f>(i) ;
// for(int j=0; j<points.cols; ++j)
// {
// out<<i<<" "<<j<<" x: "<<(*point).x<<" y: "<<(*point).y<<" z: "<<(*point).z<<endl;
// ++point;
// }
//}
waitKey(0);
return 0;
}
 | 4 | No.4 Revision |
I tried your program. I think there is a problem when you initialize the size of cm1 and cm2 (it should be [3][3]):
double cm1[3][1] = {{8.941981e+02, 0.000000e+00, 6.601406e+02}, {0.000000e+00, 8.927151e+02, 2.611004e+02}, {0.000000e+00, 0.000000e+00, 1.000000e+00}}; double cm2[3][2] = {{8.800704e+02, 0.000000e+00, 6.635881e+02 }, {0.000000e+00, 8.798504e+02, 2.690108e+02}, {0.000000e+00, 0.000000e+00, 1.000000e+00}};
It tried to visualize your result with Meshlab (rename the file to .xyz and remove the header) as I don't have for the moment PCL:

For
reprojectImageTo3D
I think that it is better to use the raw disparity instead of the normalize one. The result:

I don't think there is a problem when you reconstruct the 3D the point cloud.
Edit:
When I load the point cloud (your pcd file renamed in xyz and without the header part) into Meshlab, I get this:

and I have to move around to get the view of the first image (rotation on two axis + zoom).
Also, to obtain the point cloud in the second image, I had to manually threshold the value:
if(points.at<Vec3f>(i,j)[2] < 10) {
//Write
}
I use this tutorial to get better visualization.
The code I used:
#include <iostream>
#include <fstream>
#include <opencv2/core/core.hpp>
#include <opencv2/calib3d/calib3d.hpp>
#include <opencv2/highgui/highgui.hpp>
#include <opencv2/imgproc/imgproc.hpp>
using namespace cv;
using namespace std;
int main()
{
Mat img1, img2;
img1 = imread("left.png", CV_LOAD_IMAGE_GRAYSCALE);
img2 = imread("right.png", CV_LOAD_IMAGE_GRAYSCALE);
double cm1[3][3] = {{8.941981e+02, 0.000000e+00, 6.601406e+02}, {0.000000e+00, 8.927151e+02, 2.611004e+02}, {0.000000e+00, 0.000000e+00, 1.000000e+00}};
double cm2[3][3] = {{8.800704e+02, 0.000000e+00, 6.635881e+02 }, {0.000000e+00, 8.798504e+02, 2.690108e+02}, {0.000000e+00, 0.000000e+00, 1.000000e+00}};
double d1[1][5] = {{ -3.695739e-01, 1.726456e-01, -1.281525e-03, 1.188796e-03, -4.284730e-02}};
double d2[1][5] = {{-3.753454e-01, 1.843265e-01, -1.307069e-03, 2.190397e-03, -4.989103e-02}};
Mat CM1 (3,3, CV_64FC1, cm1);
Mat CM2 (3,3, CV_64FC1, cm2);
Mat D1(1,5, CV_64FC1, d1);
Mat D2(1,5, CV_64FC1, d2);
cout << "Calibration matrix left:\n" << CM1 << endl;
cout << "Distorstion matrix left:\n" << D1 << endl;
cout << "Calibration matrix right:\n" << CM2 << endl;
cout << "Distorstion matrix right:\n" << D2 << endl;
double r[3][3] = {{9.998381e-01, 1.610234e-02, 8.033237e-03},{-1.588968e-02, 9.995390e-01, -2.586908e-02 },{-8.446087e-03, 2.573724e-02, 9.996331e-01}};
double t[3][4] = {{ -5.706425e-01}, {8.447320e-03}, {1.235975e-02}};
Mat R (3,3, CV_64FC1, r);
Mat T (3,1, CV_64FC1, t);
//Mat R, T;
Mat R1, R2, T1, T2, Q, P1, P2;
stereoRectify(CM1, D1, CM2, D2, img1.size(), R, T, R1, R2, P1, P2, Q);
Mat map11, map12, map21, map22;
Size img_size = img1.size();
initUndistortRectifyMap(CM1, D1, R1, P1, img_size, CV_16SC2, map11, map12);
initUndistortRectifyMap(CM2, D2, R2, P2, img_size, CV_16SC2, map21, map22);
Mat img1r, img2r;
remap(img1, img1r, map11, map12, INTER_LINEAR);
remap(img2, img2r, map21, map22, INTER_LINEAR);
//img1 = img1r;
//img2 = img2r;
int sadSize = 3;
StereoSGBM sbm;
sbm.SADWindowSize = sadSize;
sbm.numberOfDisparities = 144;//144; 128
sbm.preFilterCap = 10; //63
sbm.minDisparity = 0; //-39; 0
sbm.uniquenessRatio = 10;
sbm.speckleWindowSize = 100;
sbm.speckleRange = 32;
sbm.disp12MaxDiff = 1;
sbm.fullDP = true;
sbm.P1 = sadSize*sadSize*4;
sbm.P2 = sadSize*sadSize*32;
Mat disp, disp8;
sbm(img1, img2, disp);
normalize(disp, disp8, 0, 255, CV_MINMAX, CV_8U);
Mat points, points1;
reprojectImageTo3D(disp, points, Q, true);
cvtColor(points, points1, CV_BGR2GRAY);
ofstream point_cloud_file;
point_cloud_file.open ("point_cloud.xyz");
for(int i = 0; i < points.rows; i++) {
for(int j = 0; j < points.cols; j++) {
if(points.at<Vec3f>(i,j)[2] < 10) {
point_cloud_file << points.at<Vec3f>(i,j)[0] << " " << points.at<Vec3f>(i,j)[1] << " " << points.at<Vec3f>(i,j)[2]
<< " " << static_cast<unsigned>(img1.at<uchar>(i,j)) << " " << static_cast<unsigned>(img1.at<uchar>(i,j)) << " " << static_cast<unsigned>(img1.at<uchar>(i,j)) << endl;
}
}
}
point_cloud_file.close();
imshow("Img1", img1);
imshow("Img2", img2);
imshow("points", points);
imshow("points1", points1);
//imwrite("disparity.jpg", disp8);
//imwrite("points1.jpg", points1);
//imwrite("points.jpg", points);
//for(int i=0; i<points.rows; ++i)
//{
// Point3f* point = points.ptr<Point3f>(i) ;
// for(int j=0; j<points.cols; ++j)
// {
// out<<i<<" "<<j<<" x: "<<(*point).x<<" y: "<<(*point).y<<" z: "<<(*point).z<<endl;
// ++point;
// }
//}
waitKey(0);
return 0;
}