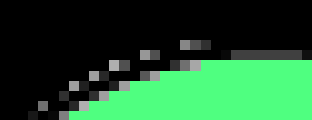This forum is disabled, please visit https://forum.opencv.org
 | 1 | initial version |
if you want to partition color data, not points, ofc. you need to adapt the Distance function:
struct ColorDist {
bool operator()(const Vec3b& a, const Vec3b &b) {
return norm(a,b) < 65; // experimental value.
}
};
// ----
Mat m=imread("part.bmp",1);
resize(m,m,Size(), 0.2, 0.2); // i'm in a hurry ;)
cerr << m.total() << " pixels." << endl;
// pack our pixels into a vector (better idea, anyone ?)
vector<Vec3f> data;
data.insert(data.begin(), m.begin<Vec3b>(), m.end<Vec3b>());
// partition labels:
vector<int> labels;
partition(data, labels, ColorDist());
cerr << labels.size() << " labels." << endl;
// count the unique ones:
set<int> clusters;
for (size_t i=0; i<labels.size(); i++)
{
clusters.insert(labels[i]);
}
cerr << clusters.size() << " clusters." << endl;
alternatively, you could just throw an integer representation of the pixel into a set, and count the uniques directly:
set<int> clusters2;
for (size_t i=0; i<m.total(); i++)
{
Vec3b v = m.at<Vec3b>(i);
int o = 7; // scale down to few bits (noise)
int c = ((v[0]>>o) << 16) | ((v[1]>>o) << 8) | (v[2]>>o);
clusters2.insert(c);
}
cerr << clusters2.size() << " clusters2." << endl;
 | 2 | No.2 Revision |
if you want to partition color data, not points, ofc. you need to adapt the Distance function:
struct ColorDist {
bool operator()(const Vec3b& a, const Vec3b &b) {
return norm(a,b) < 65; // experimental value.
}
};
// ----
Mat m=imread("part.bmp",1);
resize(m,m,Size(), 0.2, 0.2); // i'm in a hurry ;)
cerr << m.total() << " pixels." << endl;
// pack our pixels into a vector (better idea, anyone ?)
vector<Vec3f> data;
data.insert(data.begin(), m.begin<Vec3b>(), m.end<Vec3b>());
// partition labels:
vector<int> labels;
partition(data, labels, ColorDist());
cerr << labels.size() << " labels." << endl;
// count the unique ones:
set<int> clusters;
for (size_t i=0; i<labels.size(); i++)
{
clusters.insert(labels[i]);
}
cerr << clusters.size() << " clusters." << endl;
alternatively, you could just throw an integer representation of the pixel into a set, and count the uniques directly:
set<int> clusters2;
for (size_t i=0; i<m.total(); i++)
{
Vec3b v = m.at<Vec3b>(i);
int o = 7; // scale down to few bits (noise)
int c = ((v[0]>>o) << 16) | ((v[1]>>o) << 8) | (v[2]>>o);
clusters2.insert(c);
}
cerr << clusters2.size() << " clusters2." << endl;
the problem with both methods is, that your image seems to have come a long way. if you take a close look, you'll see a lot of grey, dithered (compression) artefacts around your shapes, which will be unique colors, too !
 | 3 | No.3 Revision |
if you want to partition color data, not points, ofc. you need to adapt the Distance function:
struct ColorDist {
bool operator()(const Vec3b& a, const Vec3b &b) {
return norm(a,b) < 65; // experimental value.
}
};
// ----
Mat m=imread("part.bmp",1);
resize(m,m,Size(), 0.2, 0.2); // i'm in a hurry ;)
cerr << m.total() << " pixels." << endl;
// pack our pixels into a vector (better idea, anyone ?)
vector<Vec3f> data;
data.insert(data.begin(), m.begin<Vec3b>(), m.end<Vec3b>());
// partition labels:
vector<int> labels;
partition(data, labels, ColorDist());
cerr << labels.size() << " labels." << endl;
// count the unique ones:
set<int> clusters;
for (size_t i=0; i<labels.size(); i++)
{
clusters.insert(labels[i]);
}
cerr << clusters.size() << " clusters." << endl;
alternatively, you could just throw an integer representation of the pixel into a set, and count the uniques directly:
set<int> clusters2;
for (size_t i=0; i<m.total(); i++)
{
Vec3b v = m.at<Vec3b>(i);
int o = 7; // scale down to few bits (noise)
int c = ((v[0]>>o) << 16) | ((v[1]>>o) << 8) | (v[2]>>o);
clusters2.insert(c);
}
cerr << clusters2.size() << " clusters2." << endl;
the problem with both methods is, that your image seems to have come a long way. if you take a close look, you'll see a lot of grey, dithered (compression) artefacts around your shapes, which will be unique colors, too !
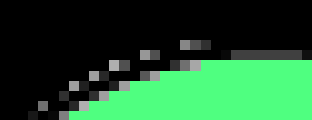
 | 4 | No.4 Revision |
if you want to partition color data, not points, ofc. you need to adapt the Distance function:
struct ColorDist {
bool operator()(const Vec3b& a, const Vec3b &b) {
return norm(a,b) < 65; // experimental value.
}
};
// ----
Mat m=imread("part.bmp",1);
resize(m,m,Size(), 0.2, 0.2); // i'm in a hurry ;)
cerr << m.total() << " pixels." << endl;
// pack our pixels into a vector (better idea, anyone ?)
vector<Vec3f> data;
data.insert(data.begin(), m.begin<Vec3b>(), m.end<Vec3b>());
// partition labels:
vector<int> labels;
int k = partition(data, labels, ColorDist());
cerr << labels.size() << " labels." << endl;
// count the unique ones:
set<int> clusters;
for (size_t i=0; i<labels.size(); i++)
{
clusters.insert(labels[i]);
}
cerr << clusters.size() k << " clusters." << endl;
alternatively, you could just throw an integer representation of the pixel into a set, map, and count the uniques directly:
set<int> clusters2;
map<int,int> clusters2; // color as key, count as value
for (size_t i=0; i<m.total(); i++)
{
Vec3b v = m.at<Vec3b>(i);
int o = 7; // scale down to few bits (noise)
int c = ((v[0]>>o) (v[0] << 16) | ((v[1]>>o) (v[1] << 8) | (v[2]>>o);
clusters2.insert(c);
(v[2]);
if (clusters2.find(c) != clusters2.end())
clusters2[c] ++;
else
clusters2[c] = 0;
}
cerr << clusters2.size() << " clusters2." << endl;
// 26 clusters.
// now, let's remove the small, noisy ones:
for (map<int,int>::iterator it=clusters2.begin(); it != clusters2.end(); )
{
if (it->second < 50) // throw away all clusters < 50 elements
it = clusters2.erase(it);
else
it++;
}
cerr << clusters2.size() << " clusters2." << endl;
// 6 clusters. ;)
the problem with both methods is, that your image seems to have come a long way. if you take a close look, you'll see a lot of grey, dithered (compression) artefacts around your shapes, which will be unique colors, too !