OpenCV Segmentation of Largest contour
Hi,
This might be a bit too "general" question, but how do I perform GRAYSCALE image segmentation and keep the largest contour? I am trying to remove background noise (i.e. labels) from breast mammograms, but I am not successful. Here is the original image:
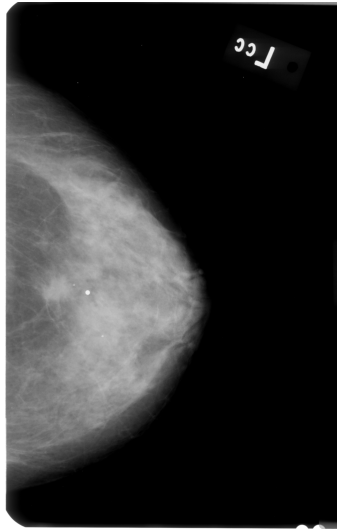
First, I applied AGCWD algorithm (based on paper "Efficient Contrast Enhancement Using Adaptive Gamma Correction With Weighting Distribution") in order to get better contrast of the image pixels, like so:
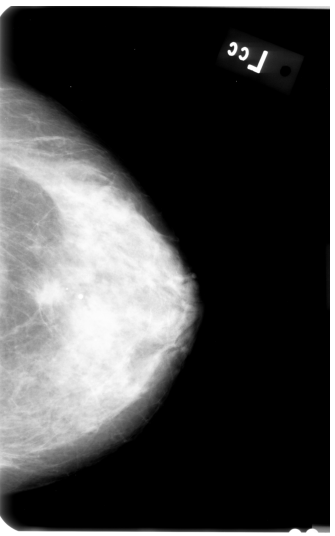
Afterwards, I tried executing following steps: Image segmentation using OpenCV's KMeans clustering algorithm:
enhanced_image_cpy = enhanced_image.copy()
reshaped_image = np.float32(enhanced_image_cpy.reshape(-1, 1))
number_of_clusters = 10
stop_criteria = (cv2.TERM_CRITERIA_EPS + cv2.TERM_CRITERIA_MAX_ITER, 100, 0.1)
ret, labels, clusters = cv2.kmeans(reshaped_image, number_of_clusters, None, stop_criteria, 10, cv2.KMEANS_RANDOM_CENTERS)
clusters = np.uint8(clusters)
Canny Edge Detection:
removed_cluster = 1
canny_image = np.copy(enhanced_image_cpy).reshape((-1, 1))
canny_image[labels.flatten() == removed_cluster] = [0]
canny_image = cv2.Canny(canny_image,100,200).reshape(enhanced_image_cpy.shape)
show_images([canny_image])
Find and Draw Contours:
initial_contours_image = np.copy(canny_image)
initial_contours_image_bgr = cv2.cvtColor(initial_contours_image, cv2.COLOR_GRAY2BGR)
_, thresh = cv2.threshold(initial_contours_image, 50, 255, 0)
contours, hierarchy = cv2.findContours(thresh, cv2.RETR_TREE, cv2.CHAIN_APPROX_SIMPLE)
cv2.drawContours(initial_contours_image_bgr, contours, -1, (255,0,0), cv2.CHAIN_APPROX_SIMPLE)
show_images([initial_contours_image_bgr])
Here is how image looks after I draw 44004 contours:
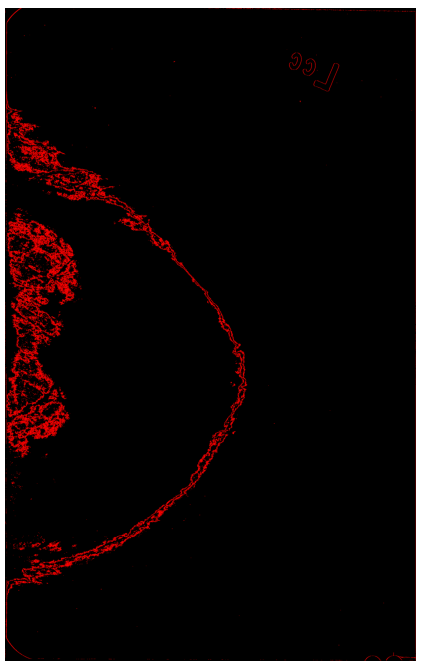
I am not sure how can I get one BIG contour, instead of 44004 small ones. Any ideas how to fix my approach, or possibly any ideas on using alternative approach to get rid of label in top right corner.
Thanks in advance!

U can't write like that
canny_image[labels.flatten() == removed_cluster] = [0]It for onlyif/elseconditionif canny_image[labels.flatten() == removed_cluster] = [0]:Found answer from stackoverflown
Easier for u to fix it. U don't need roi. Used ur code to apply this in below
ignore those comments. using a mask as an index is valid, and assigning to the result of that is also valid. I'd only question the [0] but that may be due to numpy's broadcasting rules and do the right thing.