Masking part of the image and get binary image of the masked part
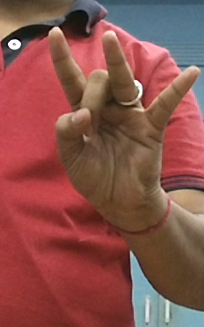
I want to mask the hand from the picture and want to get the binary image from it. Also want to measure the euclidian distance from the masked region to the entire image. I have coded so far:
# -*- coding: utf-8 -*-
"""
Created on Sun Feb 9 00:09:31 2020
@author: Shrouti
"""
import cv2
import numpy as np
from PIL import Image, ImageCms
from skimage import color
# Read input image, and create output image
src = cv2.imread("D:\SHROUTI\Testpictures\hand_gesture3.jpg")
src = cv2.resize(src, (640, 480))
# convert image to gray scale
gray = cv2.cvtColor(src, cv2.COLOR_BGR2GRAY)
# blur the image
blur = cv2.blur(gray, (3, 3))
# binary thresholding of the image
ret, thresh = cv2.threshold(blur, 200, 255, cv2.THRESH_BINARY)
# ret, thresh = cv2.threshold(gray, 127, 255,0)
# find contours
# contours, hierarchy = cv2.findContours(thresh,2,1)
contours, hierarchy = cv2.findContours(thresh, cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_NONE)
# cc
cnt = sorted(contours, key=cv2.contourArea, reverse=True)
# ROI will be object with biggest contour
mask = contours[0]
# Know the coordinates of the bounding box of the ROI
x, y, w, h = cv2.boundingRect(mask)
# Convert to Lab colourspace
Lab = color.rgb2lab(dst)
L, A, B = cv2.split(Lab)
# cv2.imshow("L_Channel", L) # For L Channel
# cv2.imshow("A_Channel", A) # For A Channel
# cv2.imshow("B_Channel", B) # For B Channel
LMean = L.mean()
AMean = A.mean()
BMean = B.mean()
cv2.waitKey(0)
cv2.destroyAllWindows()
#print(Lab)
#print(LMean)
#print(AMean)
#print(BMean)
[row,column,no_of_ColorBands]= src.shape
#make uniform images with lab colors
LStandard = LMean*np.ones([row, column], dtype = int)
AStandard = AMean*np.ones([row, column], dtype = int)
BStandard = BMean*np.ones([row, column], dtype = int)
#determine delta values
DeltaL = L-LStandard
DeltaA = A- AStandard
DeltaB = B - BStandard
DeltaE = np.sqrt(pow(DeltaA,2)+pow(DeltaB,2)+pow(DeltaL,2))
print(DeltaE)
print(mask)
#euclidian distance for only the masked region
maskedDeltaE = DeltaE*mask
add a comment

