Finding defects in the contours of a mask, derived from a complex medical image and correcting them
I have several thousand images of the lungs, taken from a CT scanner. An image looks like this.
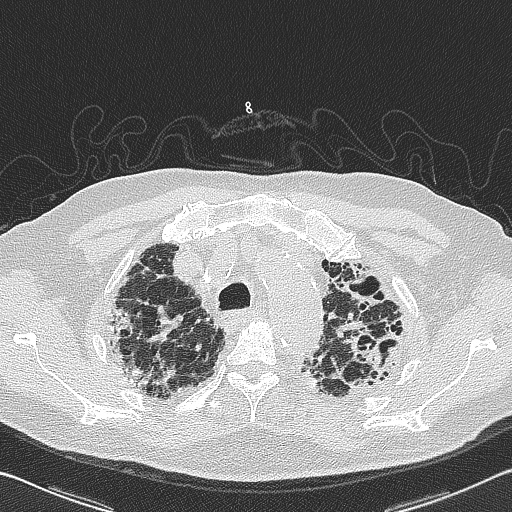
I am attempting to extract the "lungs" section from the image by creating a mask. For example:
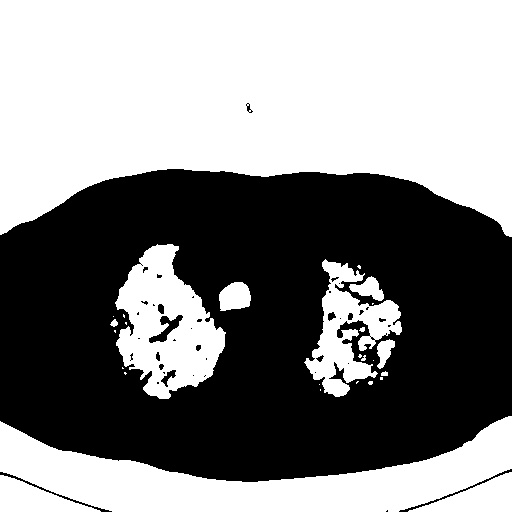
The problem is the edges of the lungs in the mask. Ideally, I could perform contour approximation to approximate the boundaries of the lungs in the mask and smooth them out so that bits weren't chopped out.
To begin with, I have tried the following code - the idea being to find the contours and then the "defects". However in this code example, defects returns None. I am new to OpenCV:
mask = cv2.imread("mask.jpg", 0)
_, contours, hierarchy = cv2.findContours(mask, 1, 2)
cnt = contours[0]
hull = cv2.convexHull(cnt, returnPoints = False)
defects = cv2.convexityDefects(cnt, hull)
for i in range(defects.shape[0]):
s,e,f,d = defects[i,0]
start = tuple(cnt[s][0])
end = tuple(cnt[e][0])
far = tuple(cnt[f][0])
cv2.line(img,start,end,[0,255,0],2)
cv2.circle(img,far,5,[0,0,255],-1)
This work is a follow on from this question on stackoverflow
https://stackoverflow.com/questions/4...
Where it was suggested I look into contour approximation. I am wondering based on what I have researched so far, if it is contour hull I am really looking for. Any suggestions would be greatly appreciated.


You're going to run into the same problem when using your own implementation of Marching Squares... I'm sorry, but what's wrong with the mask?
The edges of the mask cut out parts of the lung, because the bits of the lung on the original image at the edges are white (representing disease) - these white bits need to remain "within" the lung on the mask so that when I apply the mask to the lung image, those bits are preserved - they are important.
So basically, that particular lung slice is infested with diseased bits?
Anyway, your idea of grabbing the convex hull of the black region seems like a good solution.
You could also try running
dilate()on the mask by like 10 pixels, and thenerode()it by 10 pixels.@SLFWalsh, the
dilate()anderode()calls for Python are found here:https://docs.opencv.org/3.0-beta/doc/...
By dilating and eroding, resulting mask can be found here: https://github.com/sjhalayka/random_i...
Can you provide this code in Python?
Sorry, I edited my last comment to remove the C++ code and put in a link to some Python code that dilates and erodes.
You can also try this code:
Adjust the kernel size and iteration count to suit your needs. Also, look into
getStructuringElement(), which provides circular or cross shaped (... not just square ...) dilation/erosion elements.defects = cv2.convexityDefects(cnt, hull) is returning none value. Because of this I am not able to go further into defects.shape[0] where the error message is No shape attribute.
Could you please check this and let me know.
@Sravan, please do not post answers here, if you have a question or comment, thank you.