This forum is disabled, please visit https://forum.opencv.org
 | 1 | initial version |
First of all, I agree with @pklab: you could make your life much easier if you used a solid background
Said that, I'd take an approach using HoughCircles, as the hexagonal cells can be easily approximated
Mat image = imread("honeycomb.jpg");
Mat gray; cvtColor(image, gray, CV_BGR2GRAY);
//Trying to remove non-uniform background
vector<Mat>bgr;
split(image, bgr);
Mat binary;
threshold(bgr[0], binary, 80, 255, CV_THRESH_BINARY_INV); //thresholding blue component
Mat kernel = getStructuringElement(MORPH_RECT, Size(8, 8));
Mat kernel2 = getStructuringElement(MORPH_RECT, Size(50, 50));
morphologyEx(binary, binary, MORPH_OPEN, kernel);
morphologyEx(binary, binary, MORPH_CLOSE, kernel2);
Mat gray_filtered;
gray.copyTo(gray_filtered, binary); //copy with mask
//Retrieving circles
vector<Vec3f> circles;
HoughCircles(gray_filtered, circles, CV_HOUGH_GRADIENT, 2, 10, 100, 20, 5, 10);
for (size_t i = 0; i < circles.size(); i++)
{
Point center = Point(round(circles[i][0]), round(circles[i][1]));
int radius = round(circles[i][2]);
circle(image, center, 0, Scalar(255, 255, 255)); //draw center in white
circle(image, center, radius, Scalar(0, 0, 255), 1); //draw circle outline in red
}
imshow("Cells", image);
waitKey();
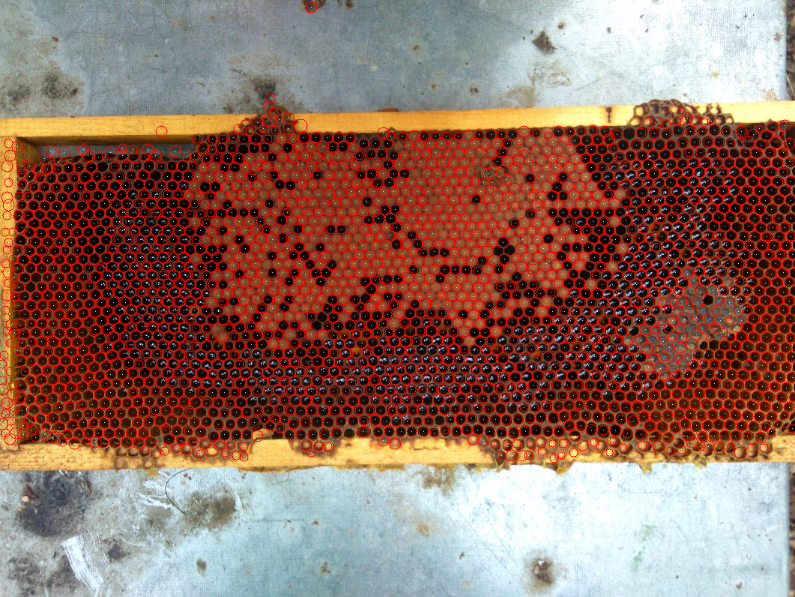
As you can see, there are still some missing cells, but a lot less than in your results. Also, there are some false positives that you can filter by defining the comb region, or by color (if that is your next step)