Identifying positive pixels after color deconvolution ignoring boundaries
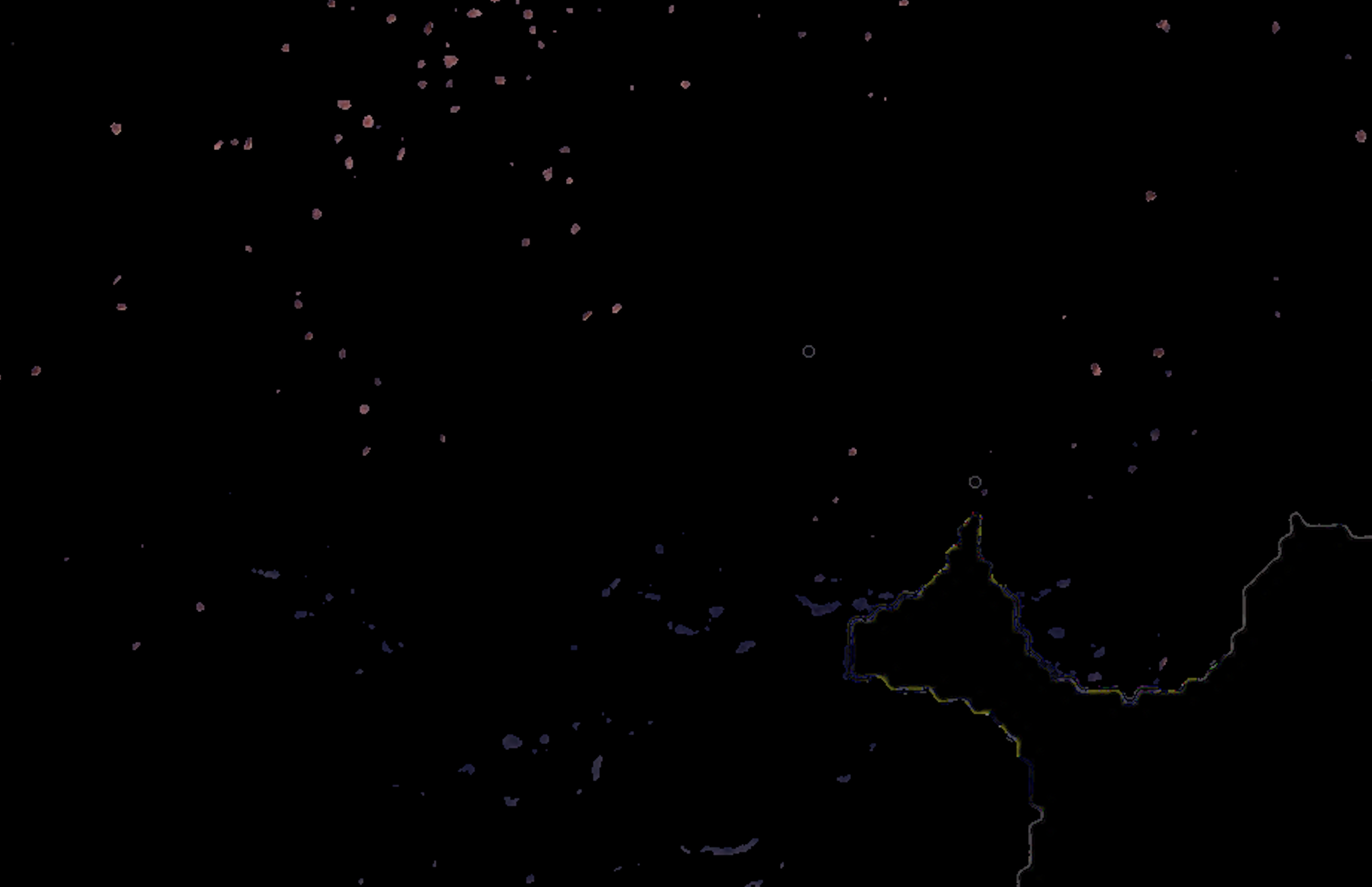
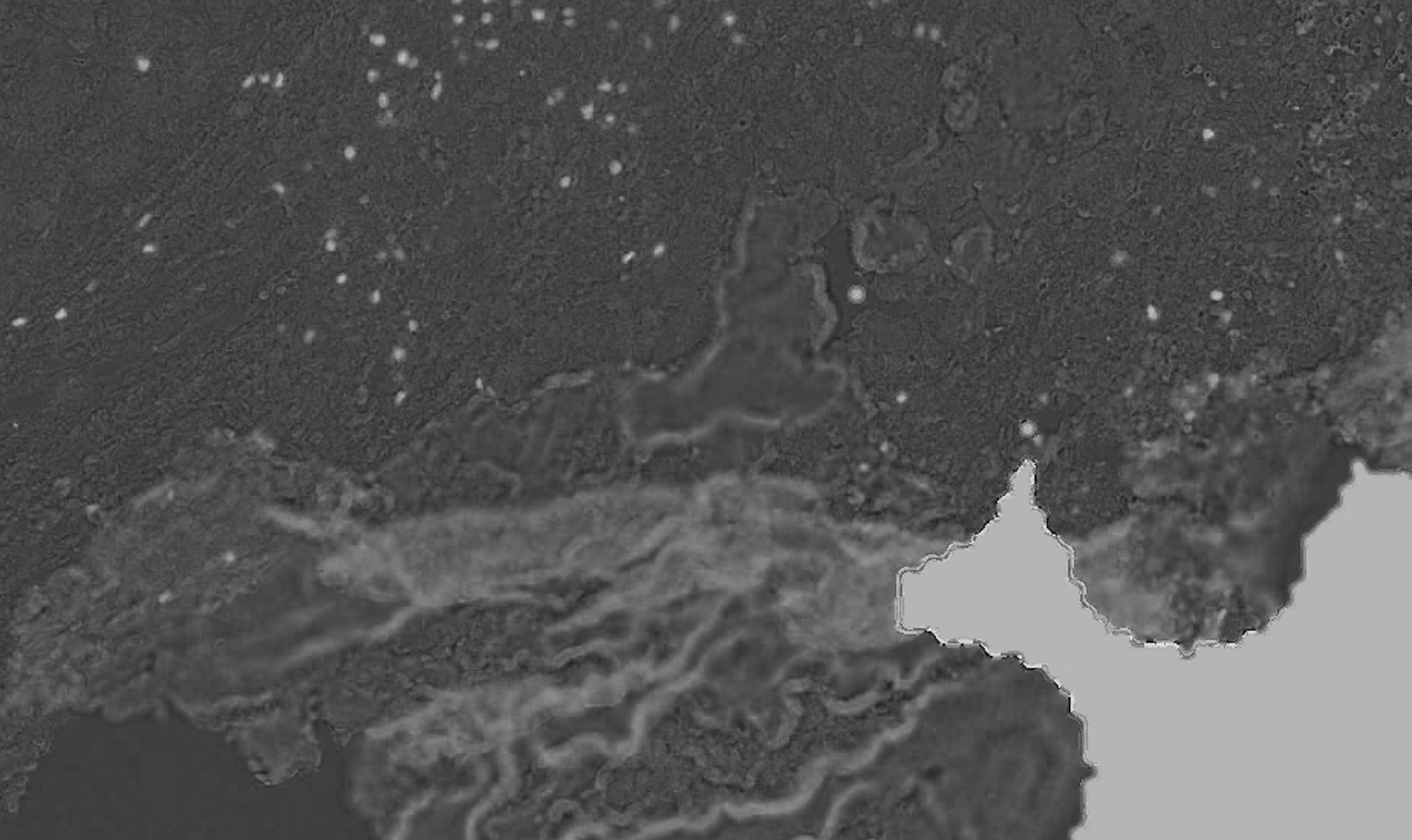
I am analyzing histology tissue images stained with a specific protein marker which I would like to identify the positive pixels for that marker. My problem is that thresholding on the image gives too much false positives which I'd like to exclude.
I am using color deconvolution (separate_stains from skimage.color) to get the AEC channel (corresponding to the red marker), separating it from the background (Hematoxylin blue color) and applying cv2 Otsu thresholding to identify the positive pixels using cv2.threshold(blur,0,255,cv2.THRESH_BINARY+cv2.THRESH_OTSU), but it is also picking up the tissue boundaries (see white lines in the example picture, sometimes it even has random colors other than white) and sometimes even non positive cells (blue regions in the example picture). It's also missing some faint positive pixels which I'd like to capture.
Overall: (1) how do I filter the false positive tissue boundaries and blue pixels? and (2) how do I adjust the Otsu thresholding to capture the faint red positives?
Thanks

Maybe think about training an object detection model.