reshape problem while training
I want to train my classifier LDA. I followed this link. but my code is crushing with memory exception and send me to this part of the code of reshape 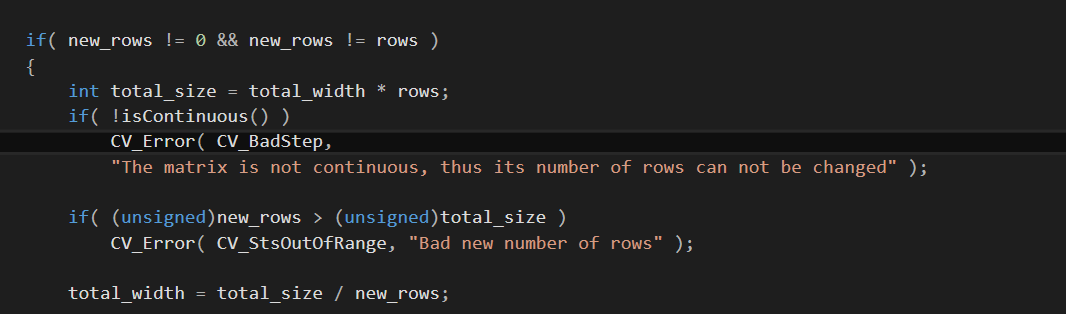 can't someone help me ?!
can't someone help me ?!
cv::Mat image1, flat_image;
cv::Mat initial_image = cv::imread("D:\mémoire\DB\to use in training\images_sorted\\image1.jpg", 0);
cv::Mat trainData = initial_image.reshape(1, 1);
////// trainData
for (int i = 2; i < 30; i++)
{
std::string filename = "D:\mémoire\DB\to use in training\images_sorted\\image";
filename = filename + std::to_string(i);
filename = filename + ".jpg";
image1 = cv::imread(filename, 0);
flat_image = image1.reshape(1, 1);
trainData.push_back(flat_image);
}

typical noob error. your image did not load (because the single backslashes in the filename) and you never checked the outcome.
DB\to usedo you vaguely remember, that\tinserts a TAB ? lol.then, no it'll NEVER work like this. the diaretdb images are 5k * 5k , you cannot use those "as is".
paper mentions SLIC clustering in LAB space, and extracting 20 features from each region ..
it's also no use, loading images as grayscale, when you're looking for yellow dots (exudates)
thanks for your replays. after applying the SLIC I extract the features so I have to give the classifier the images after applying the SLIC clustering or the original ? her an example of image they are not in the grayscale , the ground truth is color image with 2 value 0 or 255 and i'll use it for TrainLabels example . and sorry i couldn't understoood your seconde replay sorry. and you'r right i don't know how I missed the \ instead of \ -__-
I found this answer it's your i think i'll try to correct my code using it answer
have a look at the paper, again, please.
you apply ~100 SLIC clusters per image, then you gather ~20 features per cluster, stack them into a single row, and that's what you use for the classification, not the original images.
(i also wonder, how you gather the ground truth data, the supplied xml files are a total mess :\ )
you have to check:
for every imread() call you do.
just curious, you seem to be at this for quite a while now ;)
any chance, you can actually upload something to your github repos about this, so we can take a look ?
i had some issue i fixed them and I'm back + i have a professor that want to develop all the paper function in a mobile app and for the training i had to rewrite all my java code to c++ for training and the LDA classifier don't exist in opnecv java he said idk. and after wasting a lot of time he want me now to change to SVM classifier instead of LDA -__-
@break sorry for wasting your time but in the training of the SVM. the training Data i have to give to the SVM the image after preprocessing or the train Data is my features extracted from each image ? I'm confused
the features, 20 numbers per SLIC region (or from circular regions with the train data,no idea, how you handle this currently)
@break i found a good tuto of using SVM tuto but i can't find how to use my features extracted in the training + i found that some peoples say in the training Data i had to use the features extracted and not the original image.
maybe you can show, how you get your features ? actually, that part is quite the same as with LDA
this file contain my features extraction from each image . So i for each image i apply some preprocessing treatment and then i apply the SLIC superpixel (the number of regions is not the some for each image ) and for each regions i extract Min,max,mean and median form the Red,blue green saturation,gray channels as montioned in the paper. and now i'm searching how could i use them with SVM because with LDA there's lda.project where i could use my features as input
ah, thanks for the post. i actually got curious over the holidays and came up with something similar
but i see some problem coming up now: the groundtruth data has circle / polygon regions, and labels for those, while the test data is obtained via slic clustering. how to make those things fit together ? (i also don't understand the polygonregions at all there)
also, wouldn't it need to exclude the optical disk from the features ?
with LDA, you would project your train features to LDA features, and, for a 2 class system, threshold the projected values (>0 == 1)
with the SVM, you put all train features into a large train matrix, the same way as with the LDA. you just don't "project" anything, but call the predict() method onn the test feature(s) later.
i got a bit further now.
i decided to take the centroid coords from the xml files, and look up my SLIC cluster label based on the position. then i could actually start to train lda / svm.
for your question about the groundtruth. if you download the data Base you will find the image of the groundtruth png type + i applyed a threshold on the iumages of the data base to get the images with exudate beacause the different color existe in the image is the confidentiality of that region to be a exudate so the withe region are 100% to be exudate ( don't forget that the data base was marked by 4 doctors )
ah, i missed that ;) explains the 4 (slghtly different) xml files per image.
how do you handle that ?
in each xml file there's 4 image cause the database contain 3 type of lesions of diabetic retinopathy : exudate, Haemorrhage , redsmalldots and softexudate and in each image the 4 doctors marked all the lesion and we get 4 goundtruth images ;) hope you could understand me
bu the way how long it takes you to write all this code !! i'm trying to understand it ;)
@break and the optic disk is going to be deleted from the result after classification